Software
LiberTEM
The vision of LiberTEM is to make electron microscopy with high-speed detectors and complex user-defined numerical methods as easy and convenient as conventional microscopy on a phosphor screen or CCD camera. That requires exceptionally high throughput and scalability, live processing and visualization, standardized and versatile interfaces to specify a wide range of processing algorithms, and support for relevant detectors and data formats. Furthermore, it should promote Open Science and FAIR data.
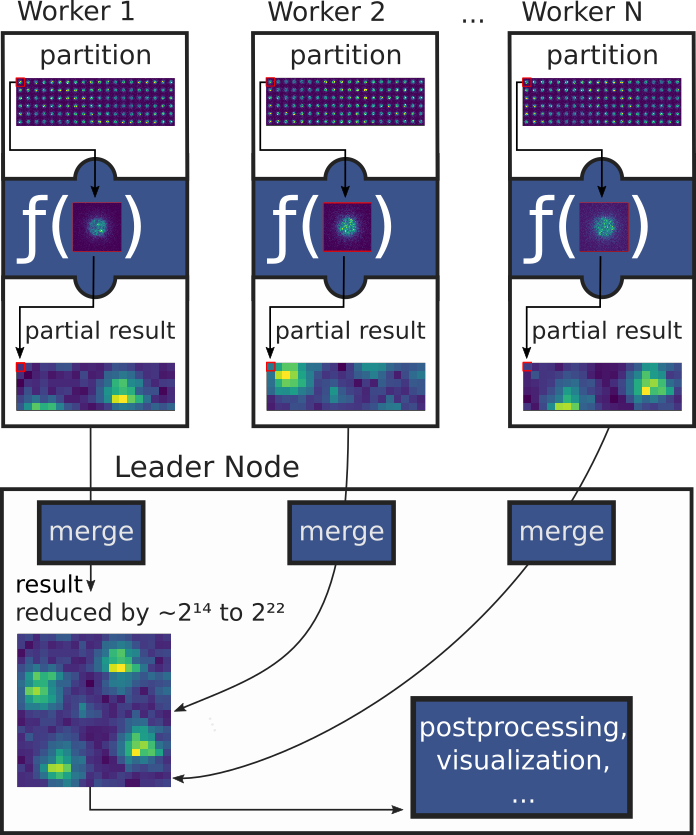
An Open Source platform for high-throughput distributed processing of large-scale binary data using a modified MapReduce programming model is created to address these challenges. Compared to Apache Spark for MapReduce processing, LiberTEM includes optimized data paths and APIs for high-performance processing of binary data, as well as relevant file format support. Compared to Dask arrays, LiberTEM allows live processing with live-updating progressive results as well as a more convenient API for non-trivial processing tasks such as ptychography or strain mapping.
Building on the MapReduce approach, the UDF interface of LiberTEM offers a standardized, versatile API to decouple the mathematical core of an algorithm from details of data source, parallelism, and use of results. That means the same implementation can be used in a wide range of modalities, including massive scaling on clusters, integration in 3rd party software and live data processing.
Furthermore, LiberTEM includes optimized back-end implementations for relevant data sources, including a wide range of file formats as well as detectors for live processing.
As an exemplary application, this infrastructure was used to implement live ptychography and center of mass analysis.
LiberTEM is Open Source and mostly licensed under GPL v3.0.